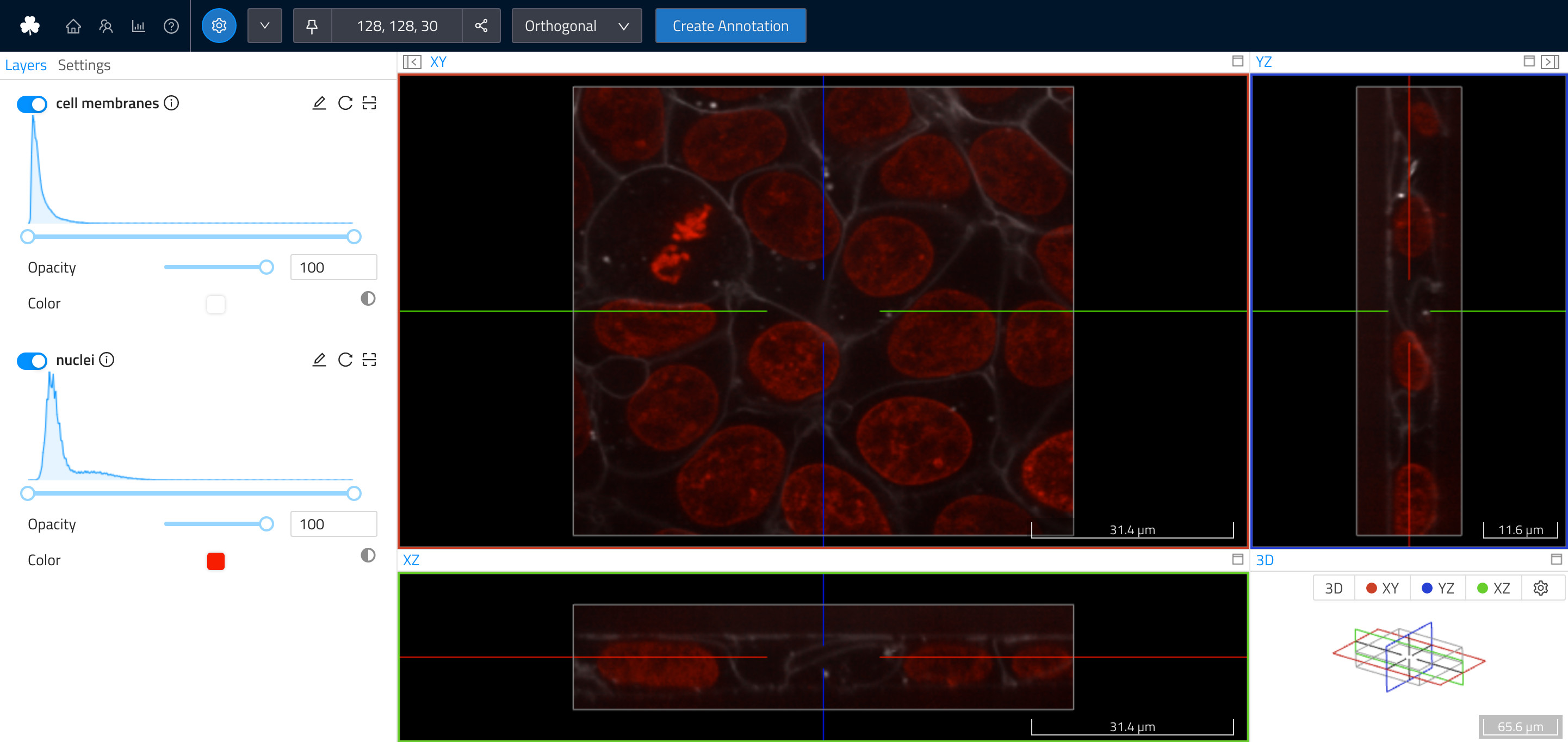
Upload Image Data¶
Loads the cell dataset, writes it into a WEBKNOSSOS Dataset and uploads it to a local webknossos instance using webknossos_context.
import numpy as np
from skimage import data
import webknossos as wk
from webknossos import COLOR_CATEGORY
from webknossos.dataset_properties import (
DatasetViewConfiguration,
LayerViewConfiguration,
)
def main() -> None:
# load your data - we use an example 3D dataset here
img = data.cells3d() # (z, c, y, x)
# make sure that the dimension of your data has the right order
# we expect the following dimensions: Channels, X, Y, Z.
img = np.transpose(img, [1, 3, 2, 0])
# choose name and voxel size (voxel_size is defined in nm)
ds = wk.Dataset("cell_dataset", voxel_size=(260, 260, 290))
ds.default_view_configuration = DatasetViewConfiguration(zoom=0.35)
# The example microscopy data has two channels
# Channel 0 contains cell membranes, channel 1 contains nuclei.
layer_membranes = ds.write_layer("cell_membranes", COLOR_CATEGORY, data=img[0, :])
layer_membranes.default_view_configuration = LayerViewConfiguration(
color=(17, 212, 17), intensity_range=(0, 16000)
)
layer_nuclei = ds.write_layer("nuclei", COLOR_CATEGORY, data=img[1, :])
layer_nuclei.default_view_configuration = LayerViewConfiguration(
color=(212, 17, 17), intensity_range=(3000, 30000)
)
remote_dataset = ds.upload()
url = remote_dataset.url
print(f"Successfully uploaded {url}")
if __name__ == "__main__":
main()
Cell Dataset uploaded to WEBKNOSSOS
- Get Help
- Community Forums
- Email Support