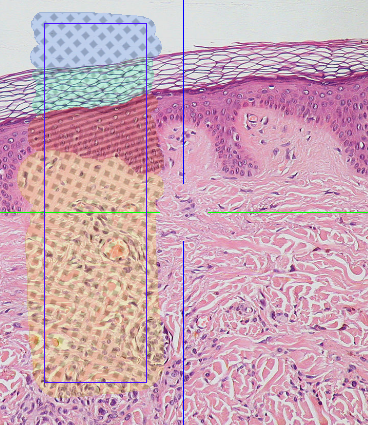
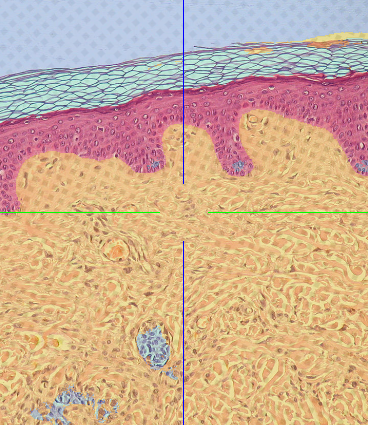
Learned Segmenter¶
This example trains a segmenter from a volume annotation and applies it to the whole dataset.
Volume annotation used for training
Result of the trained segmenter on the full dataset
It builds upon the two previous examples using the Dataset API and dataset upload. Additionally, it downloads this manual volume annotation of a subset of the skin example dataset which is used for training.
This example additionally needs the scikit-learn and pooch packages.
from functools import partial
import numpy as np
from skimage import feature
from skimage.future import TrainableSegmenter
import webknossos as wk
def main() -> None:
# We are going to use a public demo annotation for this example
annotation = wk.Annotation.download(
"https://webknossos.org/annotations/616457c2010000870032ced4"
)
# Step 1: Read the training data from the annotation and the dataset's color
# layer (the data will be streamed from WEBKNOSSOS to our local computer)
training_data_bbox = annotation.user_bounding_boxes[0] # type: ignore[index]
with wk.webknossos_context("https://webknossos.org"):
dataset = annotation.get_remote_annotation_dataset()
volume_mag_view = dataset.layers["Volume"].get_finest_mag()
mag = volume_mag_view.mag
volume_annotation_data = volume_mag_view.read(
absolute_bounding_box=training_data_bbox
)
color_mag_view = dataset.layers["color"].mags[mag]
# Step 2: Initialize a machine learning model to segment our dataset
features_func = partial(
feature.multiscale_basic_features, channel_axis=2, edges=False
)
segmenter = TrainableSegmenter(features_func=features_func)
# Step 3: Manipulate our data to fit the ML model and start training on
# data from our annotated training data bounding box
print("Starting training…")
img_data_train = color_mag_view.read(
absolute_bounding_box=training_data_bbox
) # wk data has dimensions (Channels, X, Y, Z)
# move channels to last dimension, remove z dimension to match skimage's shape
X_train = np.moveaxis(np.squeeze(img_data_train), 0, -1)
Y_train = np.squeeze(volume_annotation_data)
segmenter.fit(X_train, Y_train)
# Step 4: Use our trained model and predict a class for each pixel in the dataset
# to get a full segmentation of the data
print("Starting prediction…")
X_predict = np.moveaxis(np.squeeze(color_mag_view.read()), 0, -1)
Y_predicted = segmenter.predict(X_predict)
segmentation = Y_predicted[:, :, None] # adds z dimension
assert segmentation.max() < 256
segmentation = segmentation.astype("uint8")
# Step 5: Upload the segmentation to WEBKNOSSOS
print("Uploading segmentation…")
volume_layer = annotation.add_volume_layer("segmentation", dtype=segmentation.dtype)
with volume_layer.edit() as segmentation_layer:
segmentation_layer.bounding_box = dataset.layers["color"].bounding_box
segmentation_layer.add_mag(mag, compress=True).write(segmentation)
segmentation_layer.downsample(sampling_mode="constant_z")
url = annotation.upload()
print(f"Successfully uploaded {url}")
if __name__ == "__main__":
main()
This example is inspired by the trainable segmentation example from scikit-image.
- Get Help
- Community Forums
- Email Support